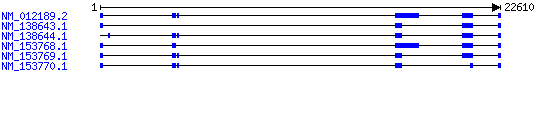Gene
Gene:
CABYR (official gene symbol)
Other symbol:
CABYR
CT family:
CT88
CT identifier:
CT88
Aliases from NCBI:
CBP86 , FSP-2 , MGC9117
Gene structure and chromosomal localization
There are five CABYR exons spanning 18.5 kb of genomic DNA. There are six human CABYR mRNAs, CABYR I–VI, including 5 splice variants involving two coding regions, coding region A (CR-A), and coding region B (CR-B) (Naaby-Hansen et al., 2002 PMID: 11820818). The six transcript variants encode five protein isoforms: CABYR-a, CABYR-b, CABYR-c, CABYR-d, and CABYR-e (two of the transcript variants encode the same isoform, CABYR-c).
Chromosome band: 18q11.2
See this gene in the UCSC Genome Browser :
See this gene in the NCBI Map Viewer Genome Browser :
Exon-intron structure and splicing variants
Splicing variants documented by RefSeqs:
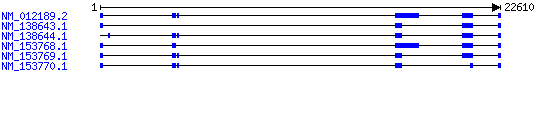
SNPs
Show
| ACCESSION |
ALLELES |
HET |
SE_HET |
FXN_CLASS |
RESIDUE |
AA_POSITION |
VALIDATED |
| rs176090 |
A/C |
0 |
0 |
coding-synonymous |
L |
17 |
YES |
| rs176090 |
A/C |
0 |
0 |
reference |
L |
17 |
YES |
| rs1049682 |
C/T |
0 |
0 |
reference |
K |
447 |
YES |
| rs1049682 |
C/T |
0 |
0 |
missense |
R |
447 |
YES |
| rs1049682 |
C/T |
0 |
0 |
reference |
K |
429 |
YES |
| rs1049682 |
C/T |
0 |
0 |
missense |
R |
429 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
missense |
A |
471 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
reference |
S |
471 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
missense |
A |
489 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
reference |
S |
489 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
reference |
A |
471 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
missense |
S |
471 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
reference |
A |
489 |
YES |
| rs1049683 |
A/C |
0.216607 |
0.24776 |
missense |
S |
489 |
YES |
| rs1049684 |
A/G |
0.496247 |
0.0431584 |
coding-synonymous |
P |
178 |
YES |
| rs1049684 |
A/G |
0.496247 |
0.0431584 |
reference |
P |
178 |
YES |
| rs1049684 |
A/G |
0.496247 |
0.0431584 |
coding-synonymous |
P |
276 |
YES |
| rs1049684 |
A/G |
0.496247 |
0.0431584 |
reference |
P |
276 |
YES |
| rs7504775 |
A/G |
0 |
0 |
coding-synonymous |
Q |
267 |
YES |
| rs7504775 |
A/G |
0 |
0 |
reference |
Q |
267 |
YES |
| rs7504775 |
A/G |
0 |
0 |
coding-synonymous |
Q |
169 |
YES |
| rs7504775 |
A/G |
0 |
0 |
reference |
Q |
169 |
YES |
| rs11552048 |
A/C |
0 |
0 |
missense |
K |
34 |
YES |
| rs11552048 |
A/C |
0 |
0 |
reference |
Q |
34 |
YES |
| rs11552049 |
A/C |
0 |
0 |
missense |
Q |
162 |
YES |
| rs11552049 |
A/C |
0 |
0 |
reference |
P |
162 |
YES |
| rs11552049 |
A/C |
0 |
0 |
missense |
Q |
64 |
YES |
| rs11552049 |
A/C |
0 |
0 |
reference |
P |
64 |
YES |
| rs11552049 |
A/C |
0 |
0 |
missense |
Q |
144 |
YES |
| rs11552049 |
A/C |
0 |
0 |
reference |
P |
144 |
YES |
| rs35118855 |
A/G |
0.00799987 |
0.062737 |
reference |
I |
167 |
YES |
| rs35118855 |
A/G |
0.00799987 |
0.062737 |
missense |
V |
167 |
YES |
| rs35118855 |
A/G |
0.00799987 |
0.062737 |
reference |
I |
185 |
YES |
| rs35118855 |
A/G |
0.00799987 |
0.062737 |
missense |
V |
185 |
YES |
| rs35239445 |
A/G |
0.0253123 |
0.109615 |
missense |
I |
290 |
NO |
| rs35239445 |
A/G |
0.0253123 |
0.109615 |
reference |
V |
290 |
NO |
| rs35239445 |
A/G |
0.0253123 |
0.109615 |
missense |
I |
272 |
NO |
| rs35239445 |
A/G |
0.0253123 |
0.109615 |
reference |
V |
272 |
NO |
| rs35554127 |
C/T |
0.0395096 |
0.134884 |
reference |
R |
340 |
YES |
| rs35554127 |
C/T |
0.0395096 |
0.134884 |
nonsense |
* |
340 |
YES |
| rs35554127 |
C/T |
0.0395096 |
0.134884 |
reference |
R |
322 |
YES |
| rs35554127 |
C/T |
0.0395096 |
0.134884 |
nonsense |
* |
322 |
YES |